Regression2 (Overfitting and Regularization)
1. Overfitting
import warnings
warnings.filterwarnings(action='ignore') # ignore errors
# 1. magic for inline plot
# 2. magic to print version
# 3. magic so that the notebook will reload external python modules
# 4. magic to enable retina (high resolution) plots
# https://gist.github.com/minrk/3301035
%matplotlib inline
%load_ext watermark
%load_ext autoreload
%autoreload
%config InlineBackend.figure_format = 'retina'
import numpy as np
import cvxpy as cvx
import matplotlib.pyplot as plt
from sklearn import linear_model
%watermark -a 'Jae H. Choi' -d -t -v -p numpy,cvxpy,matplotlib,sklearn
Jae H. Choi 2020-08-18 20:08:42
CPython 3.8.3
IPython 7.16.1
numpy 1.18.5
cvxpy 1.1.3
matplotlib 3.2.2
sklearn 0.23.1
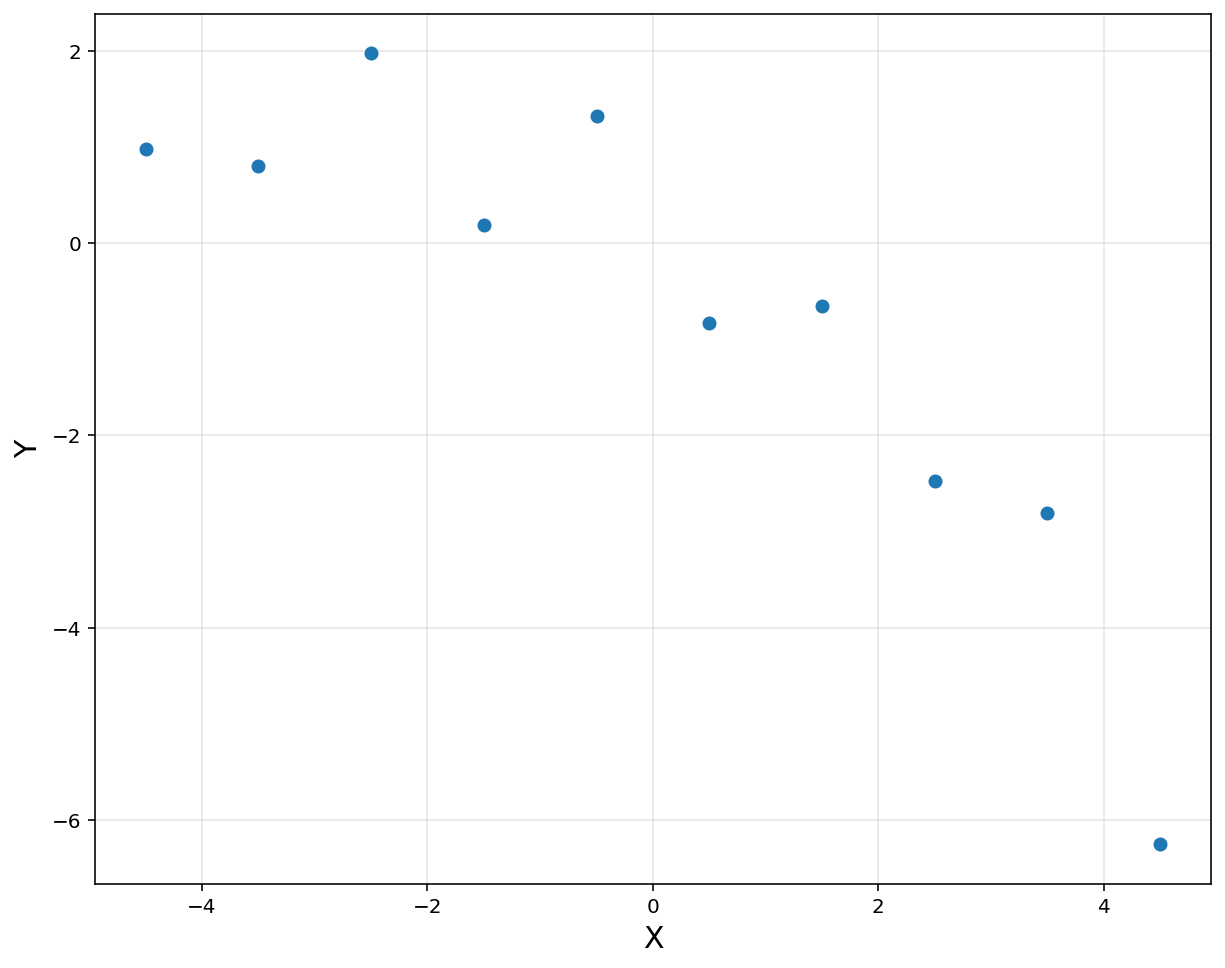
# 10 data points
n = 10
x = np.linspace(-4.5, 4.5, 10).reshape(-1, 1)
y = np.array([0.9819, 0.7973, 1.9737, 0.1838, 1.3180, -0.8361, -0.6591, -2.4701, -2.8122, -6.2512]).reshape(-1, 1)
plt.figure(figsize = (10, 8))
plt.plot(x, y, 'o', label = 'Data')
plt.xlabel('X', fontsize = 15)
plt.ylabel('Y', fontsize = 15)
plt.grid(alpha = 0.3)
plt.show()
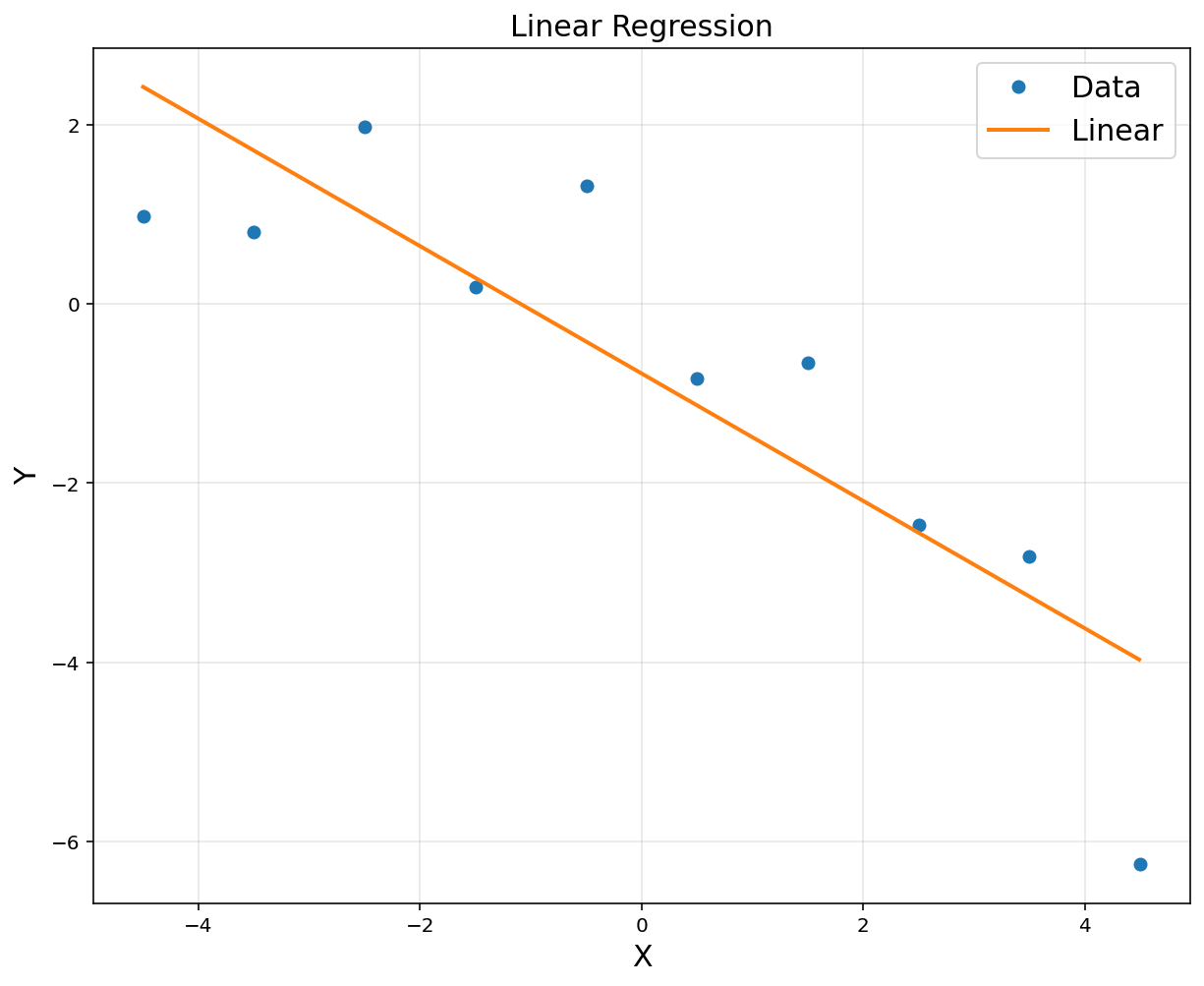
A = np.hstack([x**0, x])
A = np.asmatrix(A)
theta = (A.T*A).I*A.T*y
print(theta)
[[-0.7774 ]
[-0.71070424]]
# to plot
xp = np.arange(-4.5, 4.5, 0.01).reshape(-1, 1)
yp = theta[0,0] + theta[1,0]*xp
plt.figure(figsize = (10, 8))
plt.plot(x, y, 'o', label = 'Data')
plt.plot(xp[:,0], yp[:,0], linewidth = 2, label = 'Linear')
plt.title('Linear Regression', fontsize = 15)
plt.xlabel('X', fontsize = 15)
plt.ylabel('Y', fontsize = 15)
plt.legend(fontsize = 15)
plt.grid(alpha = 0.3)
plt.show()
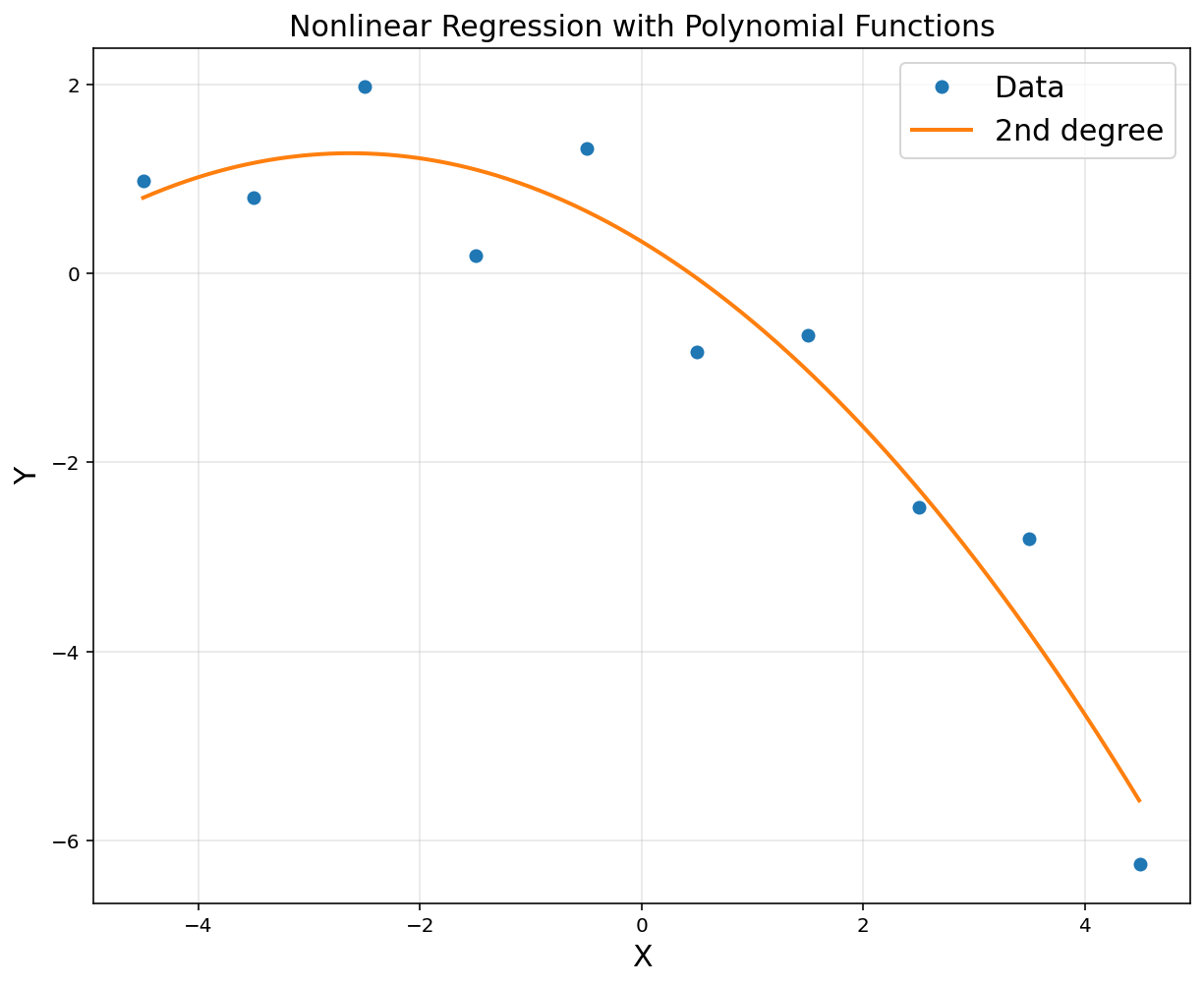
A = np.hstack([x**0, x, x**2])
A = np.asmatrix(A)
theta = (A.T*A).I*A.T*y
print(theta)
[[ 0.33669062]
[-0.71070424]
[-0.13504129]]
# to plot
xp = np.arange(-4.5, 4.5, 0.01).reshape(-1, 1)
yp = theta[0,0] + theta[1,0]*xp + theta[2,0]*xp**2
plt.figure(figsize = (10, 8))
plt.plot(x, y, 'o', label = 'Data')
plt.plot(xp[:,0], yp[:,0], linewidth = 2, label = '2nd degree')
plt.title('Nonlinear Regression with Polynomial Functions', fontsize = 15)
plt.xlabel('X', fontsize = 15)
plt.ylabel('Y', fontsize = 15)
plt.legend(fontsize = 15)
plt.grid(alpha = 0.3)
plt.show()
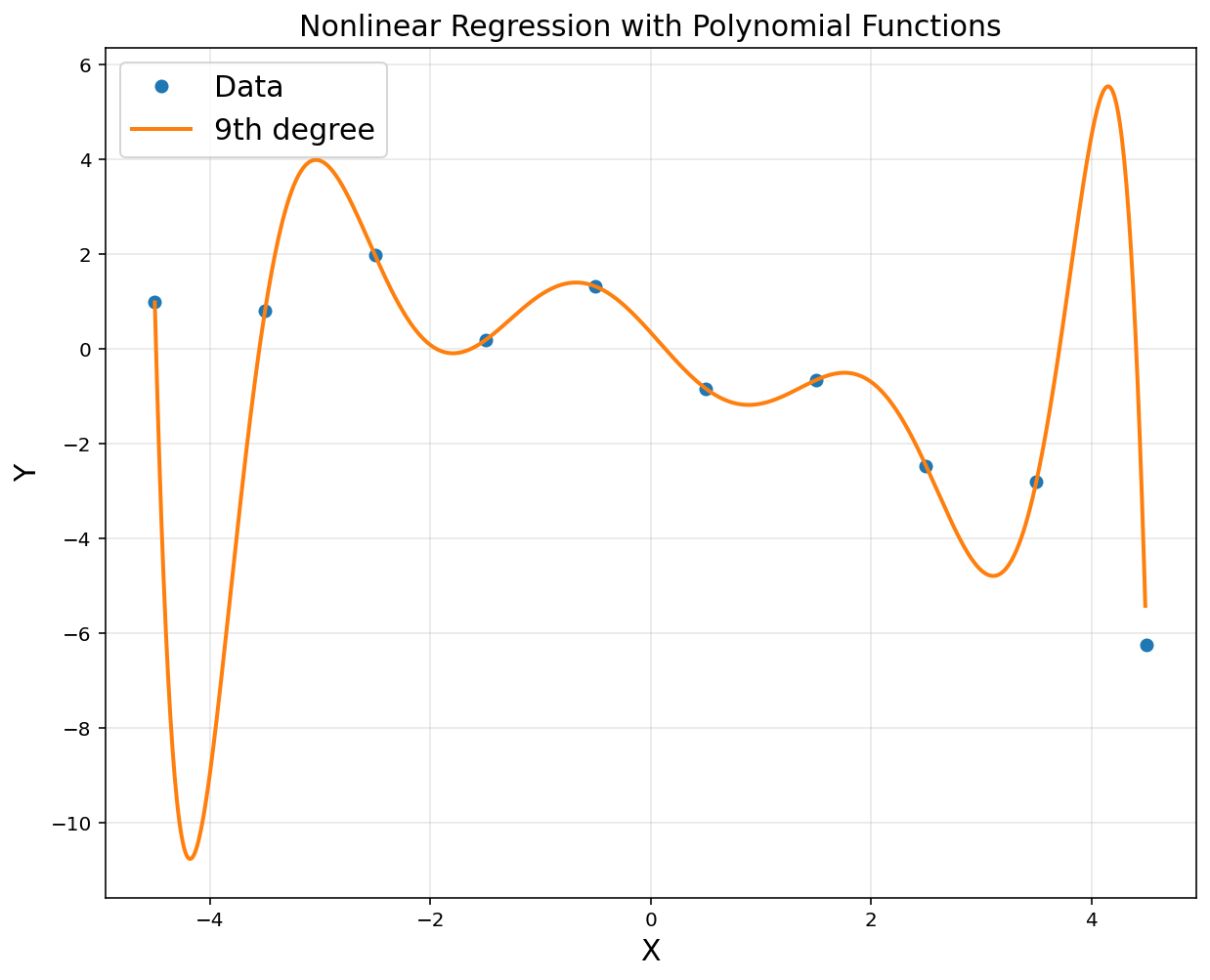
A = np.hstack([x**i for i in range(10)])
A = np.asmatrix(A)
theta = (A.T*A).I*A.T*y
print(theta)
# to plot
xp = np.arange(-4.5, 4.5, 0.01).reshape(-1, 1)
polybasis = np.hstack([xp**i for i in range(10)])
polybasis = np.asmatrix(polybasis)
yp = polybasis*theta
plt.figure(figsize = (10, 8))
plt.plot(x, y, 'o', label = 'Data')
plt.plot(xp[:,0], yp[:,0], linewidth = 2, label = '9th degree')
plt.title('Nonlinear Regression with Polynomial Functions', fontsize = 15)
plt.xlabel('X', fontsize = 15)
plt.ylabel('Y', fontsize = 15)
plt.legend(fontsize = 15)
plt.grid(alpha = 0.3)
plt.show()
[[ 3.48274701e-01]
[-2.58951123e+00]
[-4.55286474e-01]
[ 1.85022226e+00]
[ 1.06250369e-01]
[-4.43328786e-01]
[-9.25753472e-03]
[ 3.63088178e-02]
[ 2.35143849e-04]
[-9.24099978e-04]]
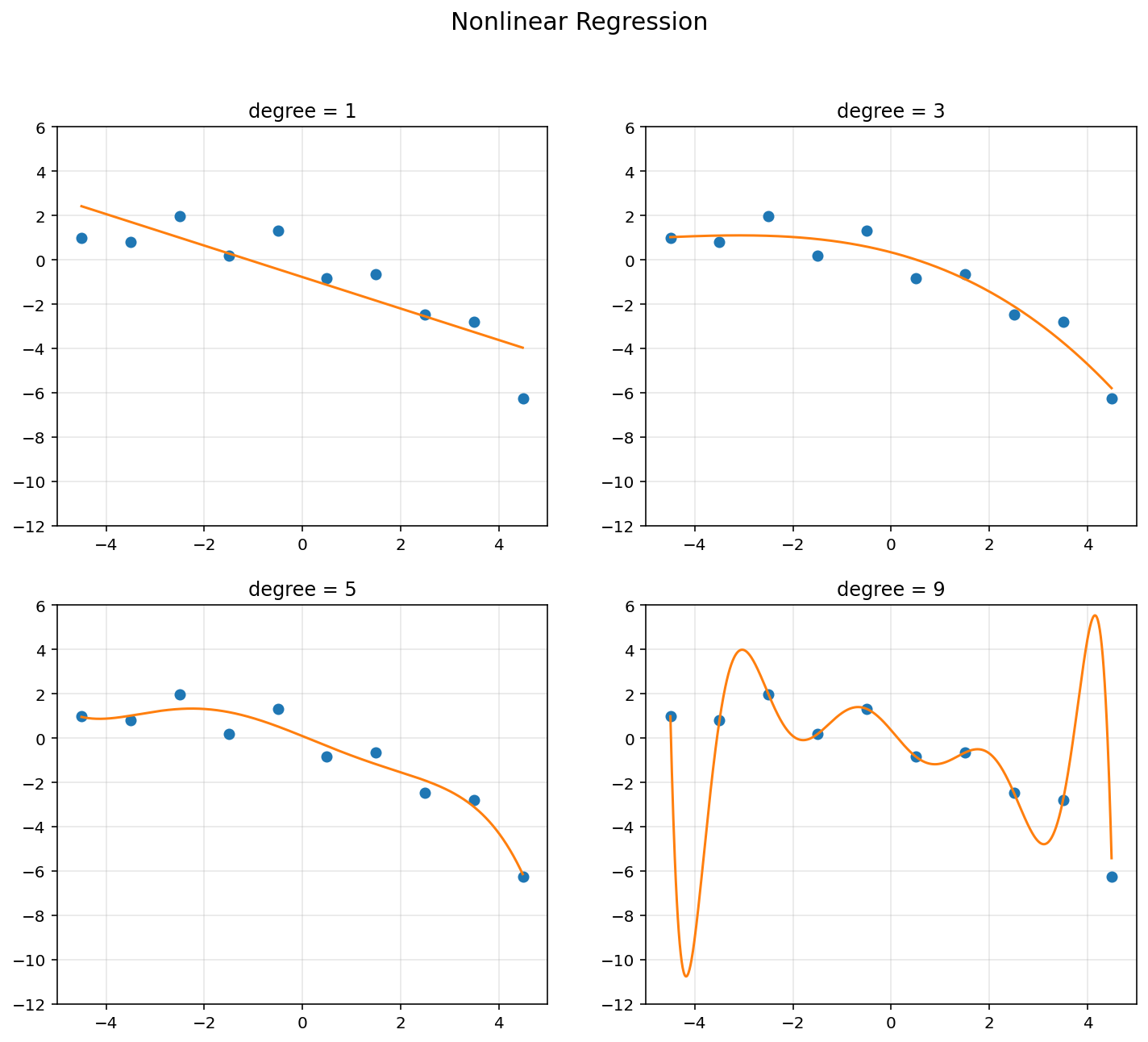
d = [1, 3, 5, 9]
RSS = []
plt.figure(figsize = (12, 10))
plt.suptitle('Nonlinear Regression', fontsize = 15)
for k in range(4):
A = np.hstack([x**i for i in range(d[k]+1)])
polybasis = np.hstack([xp**i for i in range(d[k]+1)])
A = np.asmatrix(A)
polybasis = np.asmatrix(polybasis)
theta = (A.T*A).I*A.T*y
yp = polybasis*theta
RSS.append(np.linalg.norm(y - A*theta, 2)**2)
plt.subplot(2, 2, k+1)
plt.plot(x, y, 'o')
plt.plot(xp, yp)
plt.axis([-5, 5, -12, 6])
plt.title('degree = {}'.format(d[k]))
plt.grid(alpha=0.3)
plt.show()
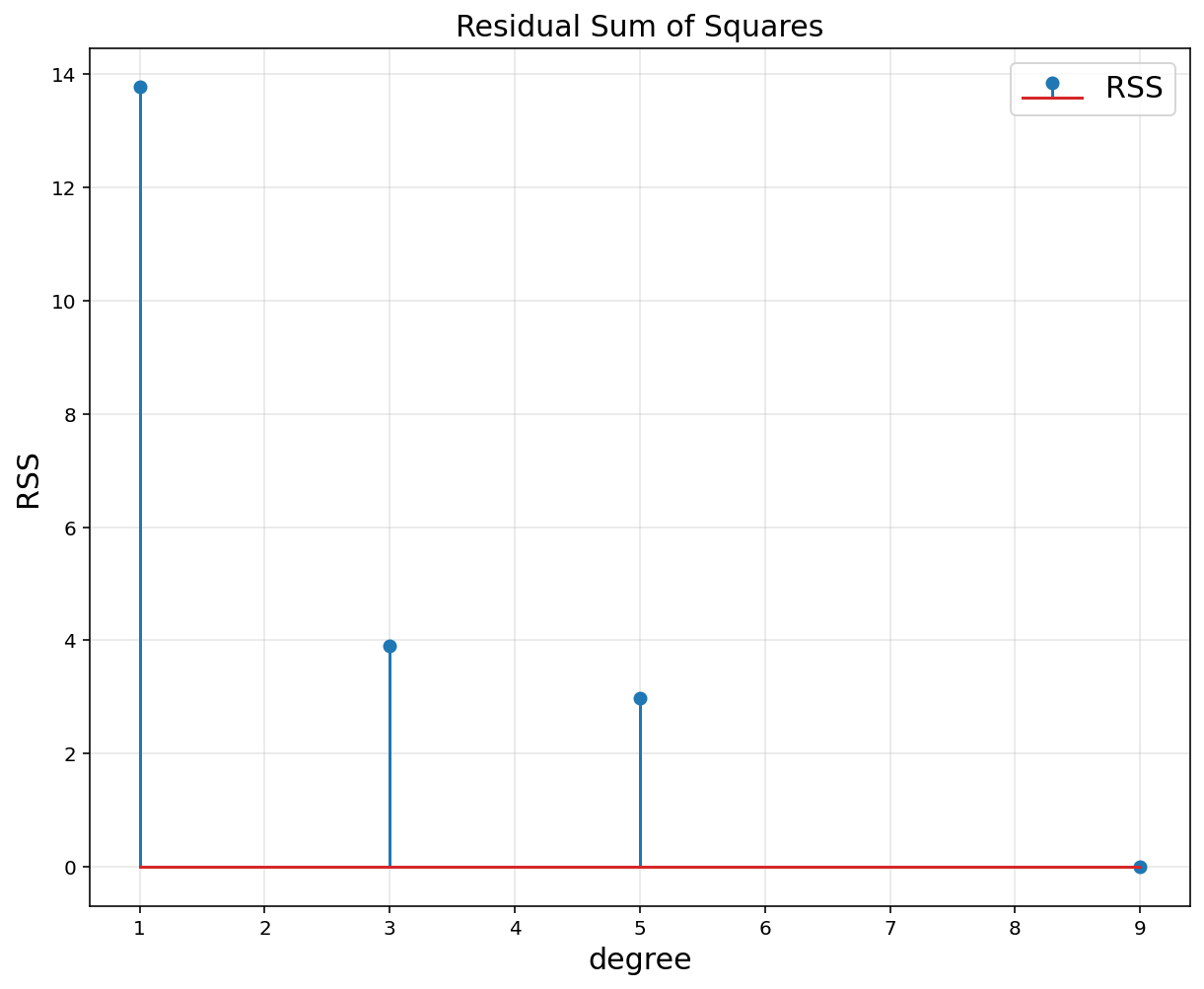
plt.figure(figsize = (10, 8))
plt.stem(d, RSS, label = 'RSS')
plt.title('Residual Sum of Squares', fontsize = 15)
plt.xlabel('degree', fontsize = 15)
plt.ylabel('RSS', fontsize = 15)
plt.legend(fontsize = 15)
plt.grid(alpha = 0.3)
plt.show()
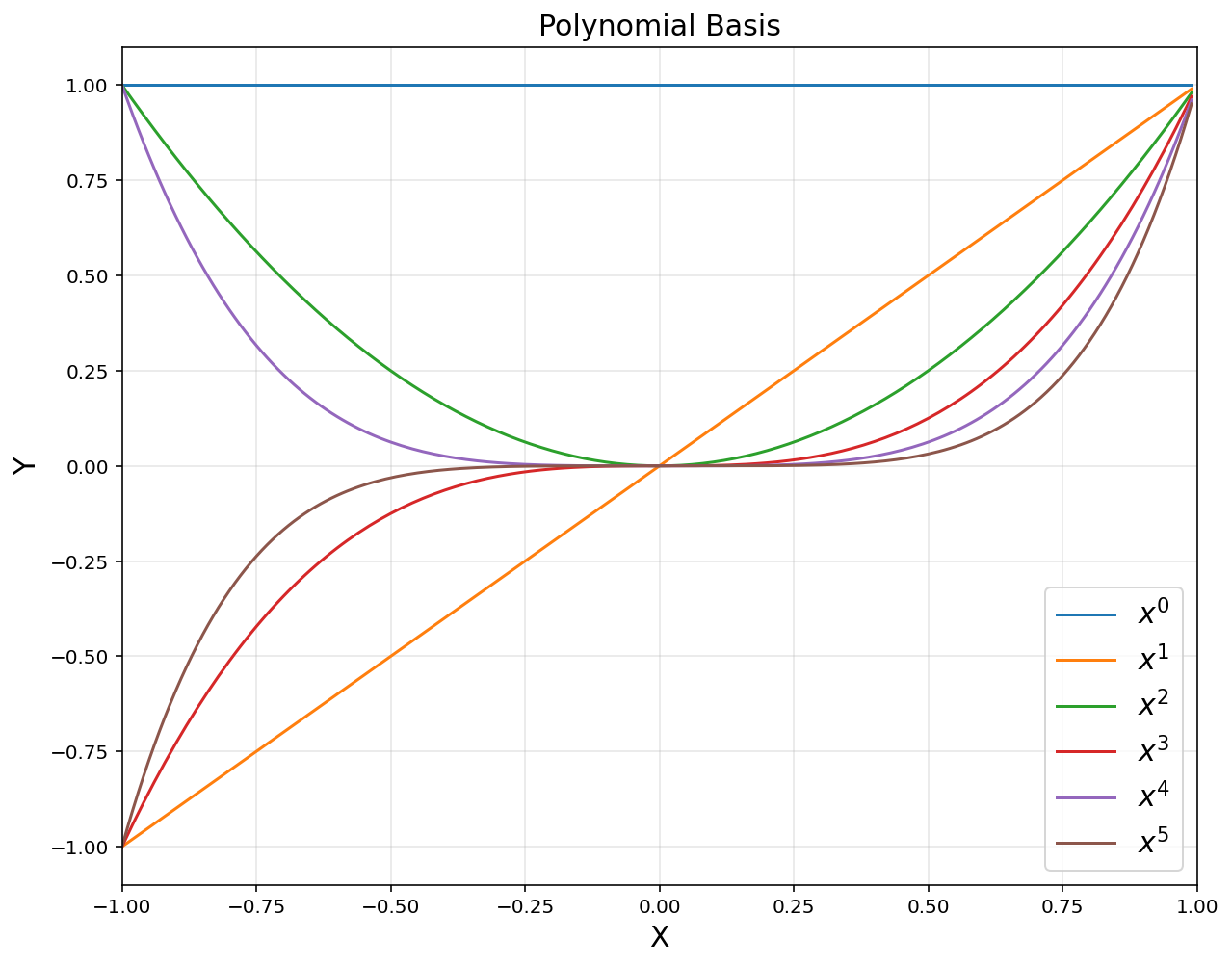
2. Linear Basis Function Models
- Construct explicit feature vectors
- Consider linear combinations of fixed nonlinear functions of the input variables, of the form
1) Polynomial functions
\[b_i(x) = x^i, \quad i = 0,\cdots,d\]xp = np.arange(-1, 1, 0.01).reshape(-1, 1)
polybasis = np.hstack([xp**i for i in range(6)])
plt.figure(figsize = (10, 8))
for i in range(6):
plt.plot(xp, polybasis[:,i], label = '$$x^{}$$'.format(i))
plt.title('Polynomial Basis', fontsize = 15)
plt.xlabel('X', fontsize = 15)
plt.ylabel('Y', fontsize = 15)
plt.axis([-1, 1, -1.1, 1.1])
plt.grid(alpha = 0.3)
plt.legend(fontsize = 15)
plt.show()
2) RBF functions With bandwidth \(\sigma\) and \(k\) RBF centers \(\mu_i \in \mathbb{R}^n\)
\[b_i(x) = \exp \left( - \frac{\lVert x-\mu_i \rVert^2}{2\sigma^2}\right)\]d = 9
u = np.linspace(-1, 1, d)
sigma = 0.2
rbfbasis = np.hstack([np.exp(-(xp-u[i])**2/(2*sigma**2)) for i in range(d)])
plt.figure(figsize = (10, 8))
for i in range(d):
plt.plot(xp, rbfbasis[:,i], label='$$\mu = {}$$'.format(u[i]))
plt.title('RBF basis', fontsize = 15)
plt.xlabel('X', fontsize = 15)
plt.ylabel('Y', fontsize = 15)
plt.axis([-1, 1, -0.1, 1.1])
plt.legend(loc = 'lower right', fontsize = 15)
plt.grid(alpha = 0.3)
plt.show()
-
With many features, our prediction function becomes very expressive
-
Can lead to overfitting (low error on input data points, but high error nearby)
xp = np.arange(-4.5, 4.5, 0.01).reshape(-1, 1)
d = 10
u = np.linspace(-4.5, 4.5, d)
sigma = 0.2
A = np.hstack([np.exp(-(x-u[i])**2/(2*sigma**2)) for i in range(d)])
rbfbasis = np.hstack([np.exp(-(xp-u[i])**2/(2*sigma**2)) for i in range(d)])
A = np.asmatrix(A)
rbfbasis = np.asmatrix(rbfbasis)
theta = (A.T*A).I*A.T*y
yp = rbfbasis*theta
plt.figure(figsize = (10, 8))
plt.plot(x, y, 'o', label = 'Data')
plt.plot(xp, yp, label = 'Overfitted')
plt.title('(Overfitted) Regression with RBF basis', fontsize = 15)
plt.xlabel('X', fontsize = 15)
plt.ylabel('Y', fontsize = 15)
plt.grid(alpha = 0.3)
plt.legend(fontsize = 15)
plt.show()
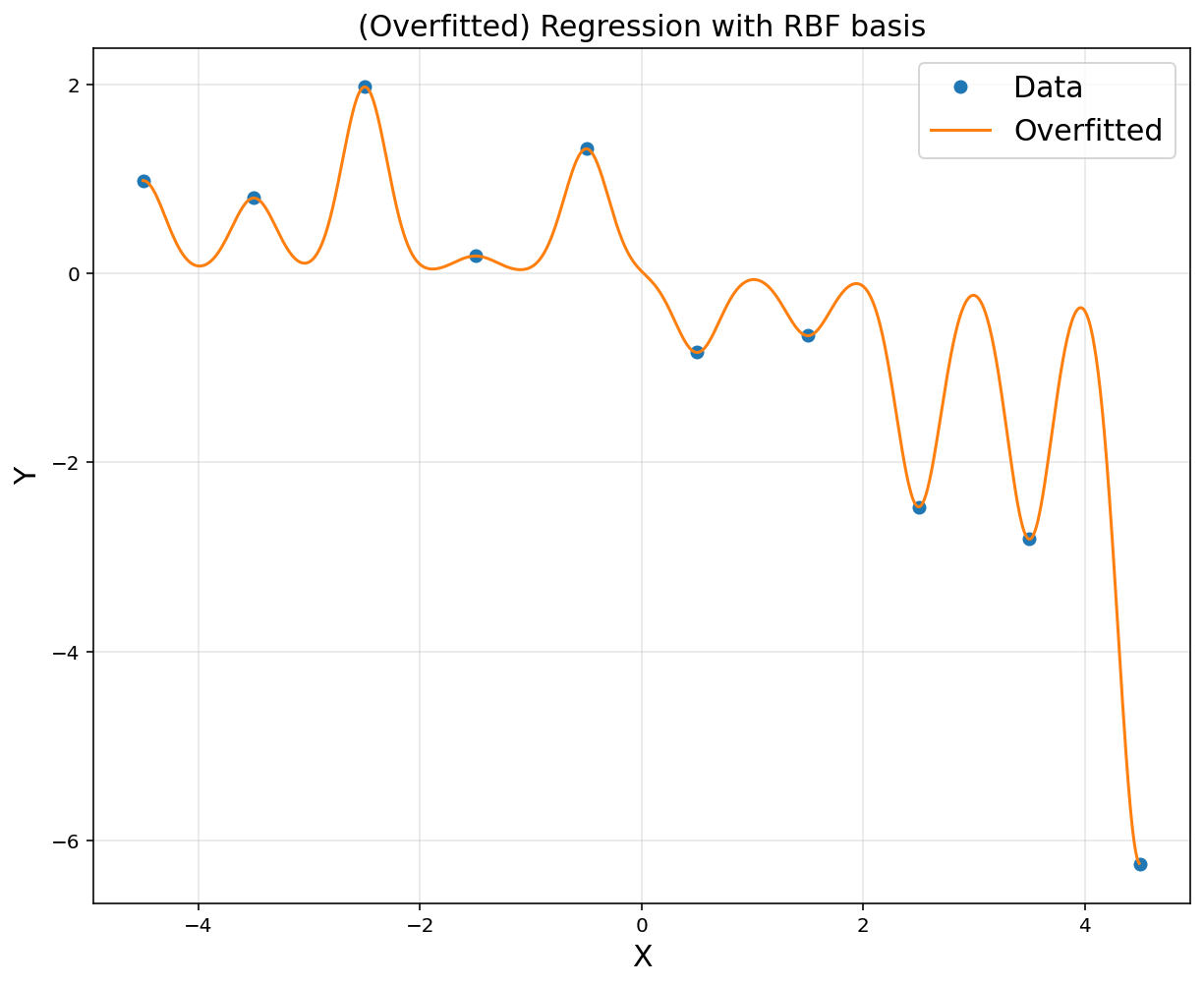
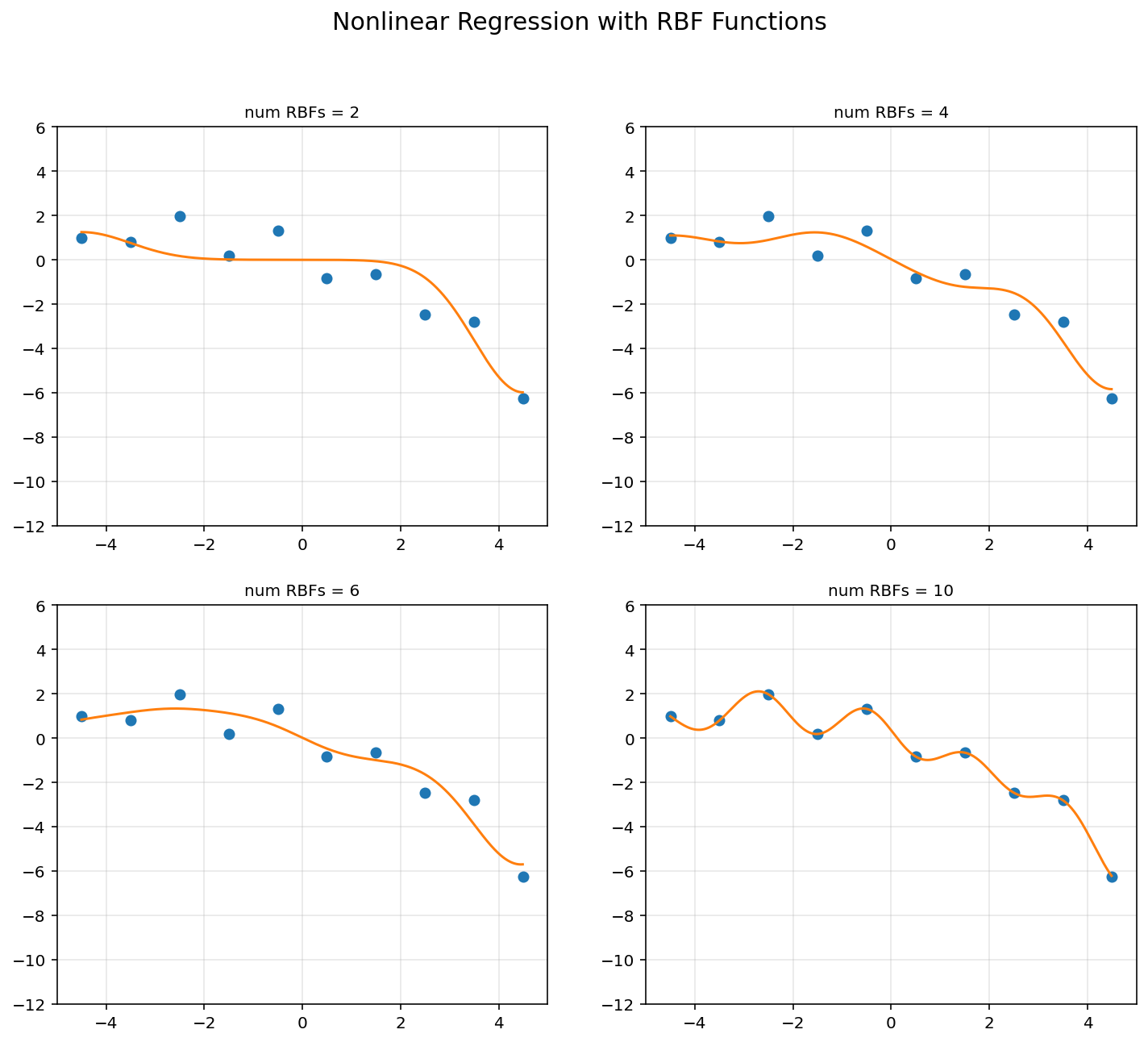
d = [2, 4, 6, 10]
sigma = 1
plt.figure(figsize = (12, 10))
for k in range(4):
u = np.linspace(-4.5, 4.5, d[k])
A = np.hstack([np.exp(-(x-u[i])**2/(2*sigma**2)) for i in range(d[k])])
rbfbasis = np.hstack([np.exp(-(xp-u[i])**2/(2*sigma**2)) for i in range(d[k])])
A = np.asmatrix(A)
rbfbasis = np.asmatrix(rbfbasis)
theta = (A.T*A).I*A.T*y
yp = rbfbasis*theta
plt.subplot(2, 2, k+1)
plt.plot(x, y, 'o')
plt.plot(xp, yp)
plt.axis([-5, 5, -12, 6])
plt.title('num RBFs = {}'.format(d[k]), fontsize = 10)
plt.grid(alpha = 0.3)
plt.suptitle('Nonlinear Regression with RBF Functions', fontsize = 15)
plt.show()
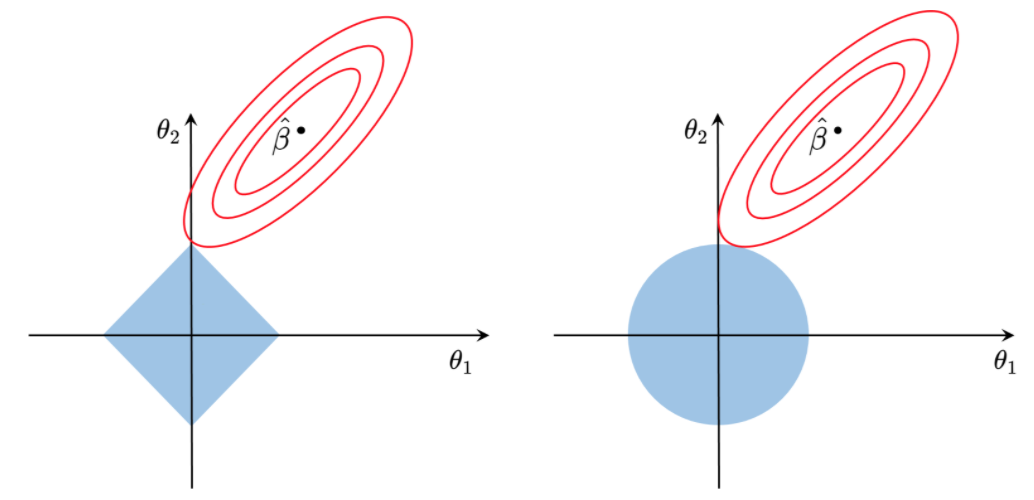
3. Regularization (Shrinkage Methods)
Often, overfitting associated with very large estimated parameters \(\theta\) We want to balance
-
how well function fits data
-
magnitude of coefficients
where \(RSS(\theta) = \lVert \Phi\theta - y \rVert^2_2\), ( = Rresidual Sum of Squares) and \(\lambda\) is a tuning parameter to be determined separately
-
the second term, \(\lambda \cdot \lVert \theta \rVert_2^2\), called a shrinkage penalty, is small when \(\theta_1, \cdots,\theta_d\) are close to zeros, and so it has the effect of shrinking the estimates of \(\theta_j\) towards zero
-
The tuning parameter \(\lambda\) serves to control the relative impact of these two terms on the regression coefficient estimates
-
known as a ridge regression
-
Example: start from rich representation.
# CVXPY code: import cvxpy as cvx
d = 10
u = np.linspace(-4.5, 4.5, d)
sigma = 1
# power(expr, p)
A = np.hstack([np.exp(-(x-u[i])**2/(2*sigma**2)) for i in range(d)])
rbfbasis = np.hstack([np.exp(-(xp-u[i])**2/(2*sigma**2)) for i in range(d)])
A = np.asmatrix(A)
rbfbasis = np.asmatrix(rbfbasis)
theta = cvx.Variable([d, 1])
obj = cvx.Minimize(cvx.sum_squares(A@theta-y))
prob = cvx.Problem(obj).solve()
yp = rbfbasis*theta.value
plt.figure(figsize = (10, 8))
plt.plot(x, y, 'o', label = 'Data')
plt.plot(xp, yp, label = 'Overfitted')
plt.title('(Overfitted) Regression', fontsize = 15)
plt.xlabel('X', fontsize = 15)
plt.ylabel('Y', fontsize = 15)
plt.axis([-5, 5, -12, 6])
plt.legend(fontsize = 15)
plt.grid(alpha = 0.3)
plt.show()
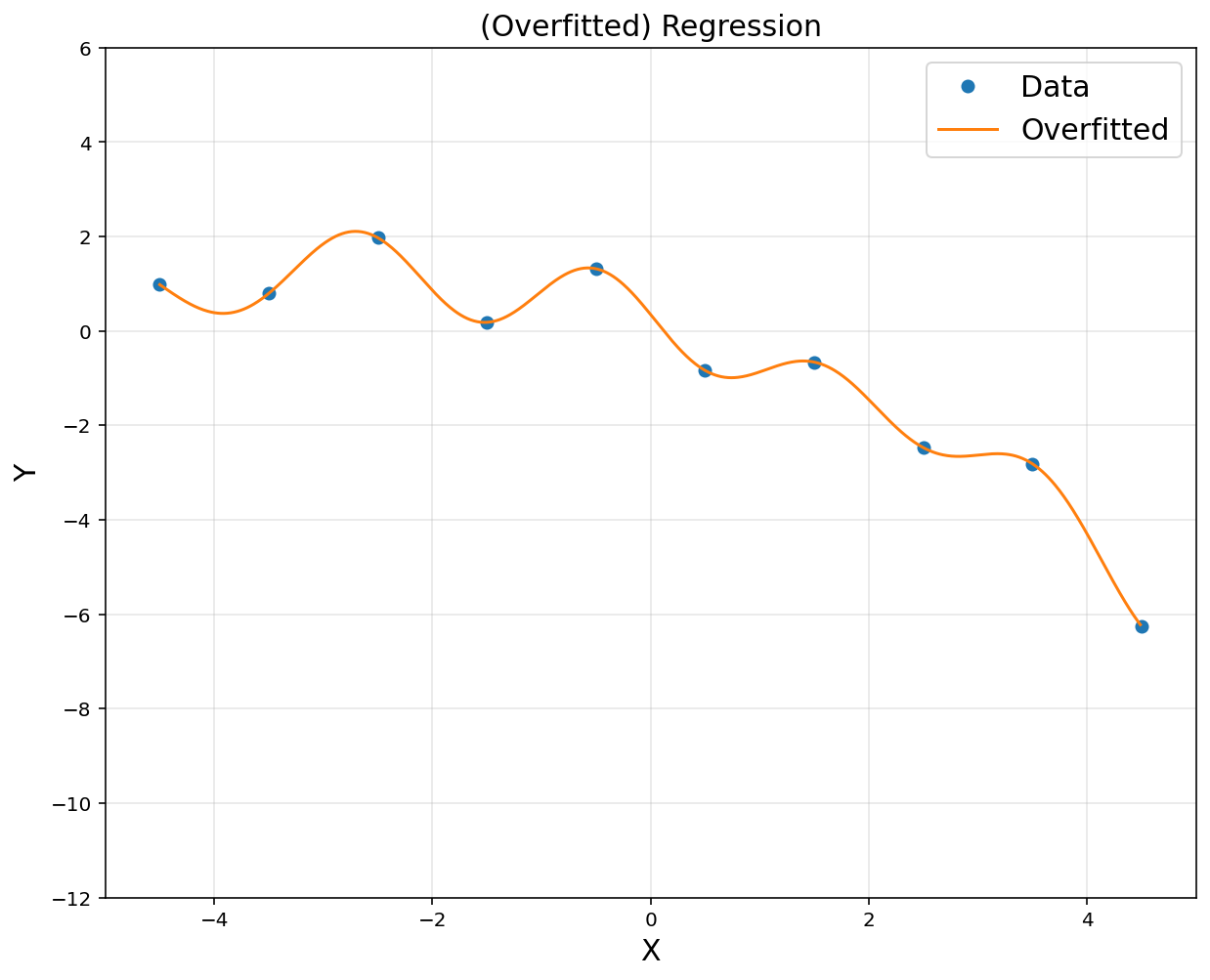
- Start from rich representation. Then, regularize coefficients \(\theta\)
# ridge regression
lamb = 0.1
theta = cvx.Variable([d, 1])
obj = cvx.Minimize(cvx.sum_squares(A@theta - y) + lamb*cvx.sum_squares(theta))
prob = cvx.Problem(obj).solve()
yp = rbfbasis*theta.value
plt.figure(figsize = (10, 8))
plt.plot(x, y, 'o', label = 'Data')
plt.plot(xp, yp, label = 'Ridge')
plt.title('Ridge Regularization (L2)', fontsize = 15)
plt.xlabel('X', fontsize = 15)
plt.ylabel('Y', fontsize = 15)
plt.axis([-5, 5, -12, 6])
plt.legend(fontsize = 15)
plt.grid(alpha = 0.3)
plt.show()
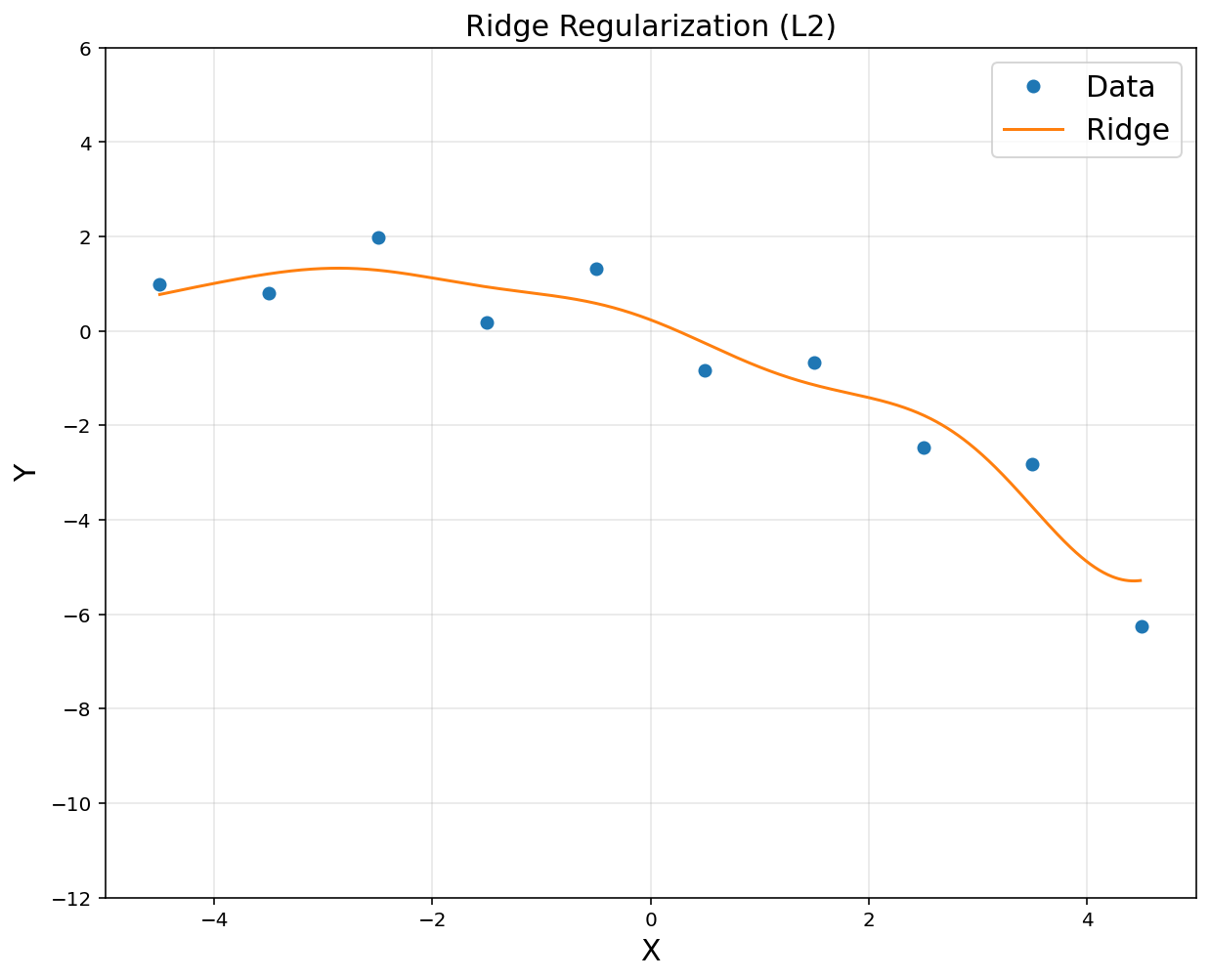
# Regulization (= ridge nonlinear regression) encourages small weights, but not exactly 0
plt.figure(figsize = (10, 8))
plt.title(r'Ridge: magnitude of $$\theta$$', fontsize = 15)
plt.xlabel(r'$$\theta$$', fontsize = 15)
plt.ylabel('magnitude', fontsize = 15)
plt.stem(np.linspace(1, 10, 10).reshape(-1, 1), theta.value)
plt.xlim([0.5, 10.5])
plt.ylim([-5, 5])
plt.grid(alpha = 0.3)
plt.show()
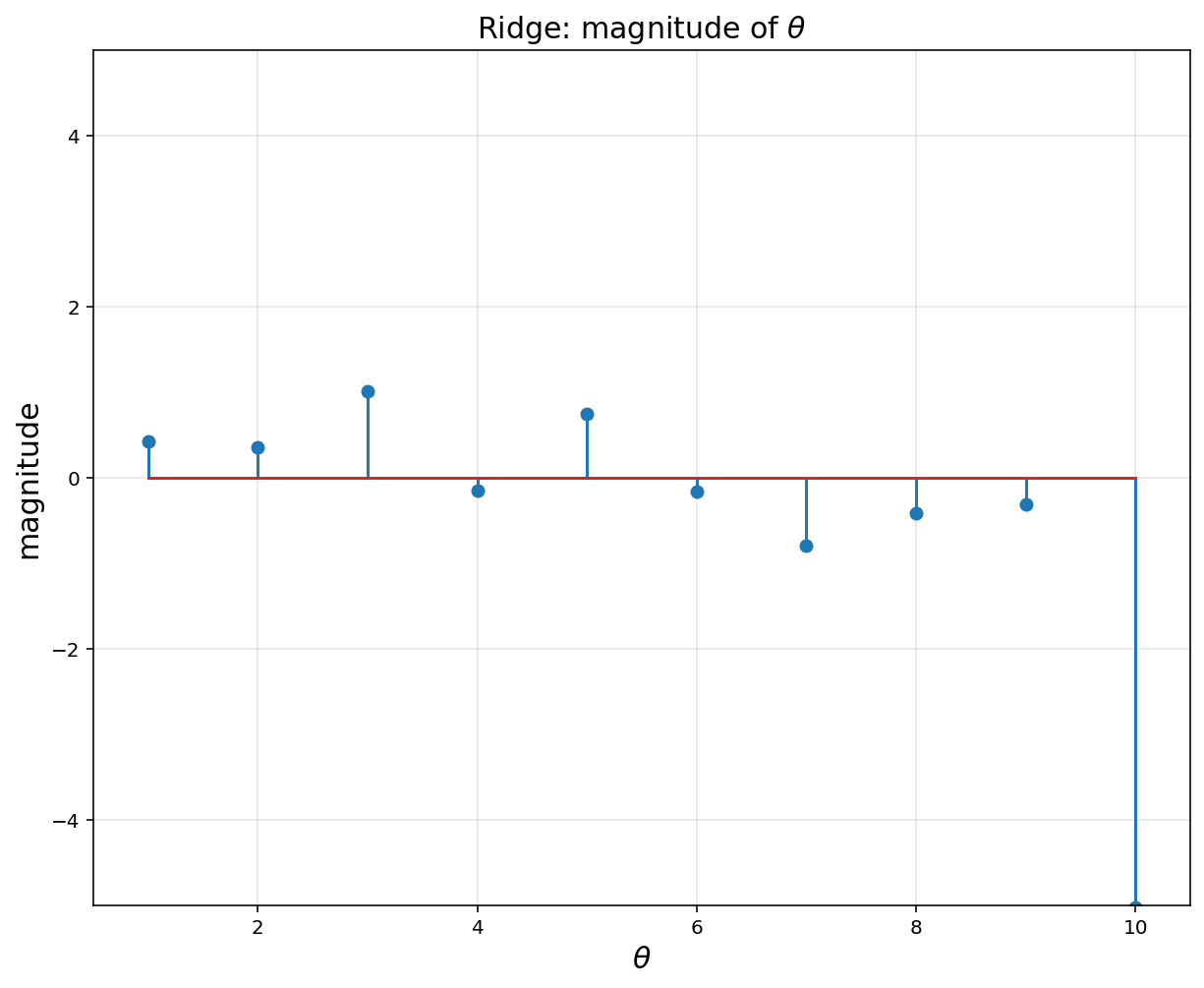
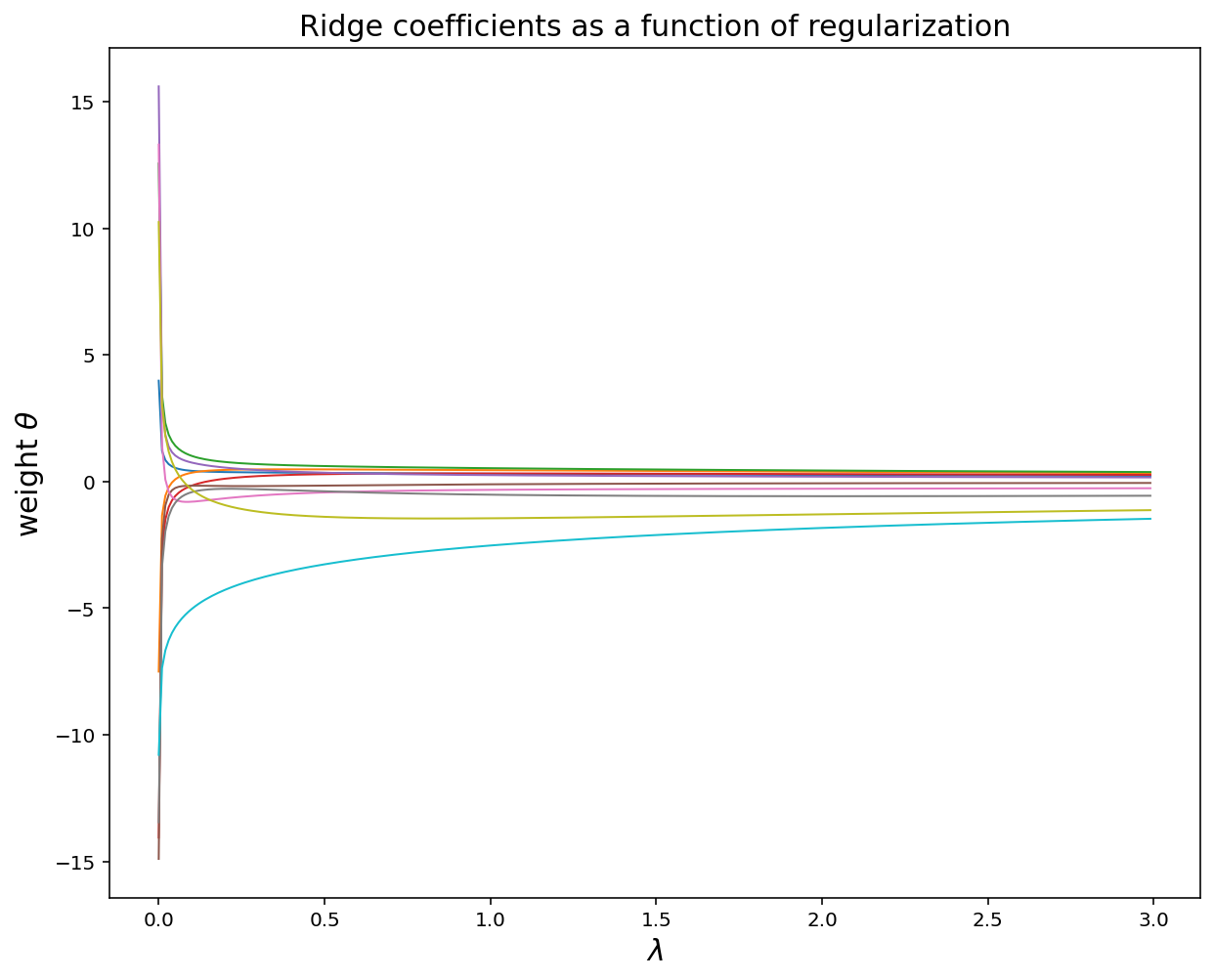
lamb = np.arange(0,3,0.01)
theta_record = []
for k in lamb:
theta = cvx.Variable([d, 1])
obj = cvx.Minimize(cvx.sum_squares(A@theta - y) + k*cvx.sum_squares(theta))
prob = cvx.Problem(obj).solve()
theta_record.append(np.ravel(theta.value))
plt.figure(figsize = (10, 8))
plt.plot(lamb, theta_record, linewidth = 1)
plt.title('Ridge coefficients as a function of regularization', fontsize = 15)
plt.xlabel('$$\lambda$$', fontsize = 15)
plt.ylabel(r'weight $$\theta$$', fontsize = 15)
plt.show()
4. Sparsity for Feature Selection using LASSO
-
Least Squares with a penalty on the \(l_1\) norm of the parameters
-
Start with full model (all possible features)
- ‘Shrink’ some coefficients exactly to 0
- i.e., knock out certain features
- the \(l_1\) penalty has the effect of forcing some of the coefficient estimates to be exactly equal to zero
- Non-zero coefficients indicate ‘selected’ features
Try this cost instead of ridge…
\[\begin{align*} \text{Total cost } = \; \ \underbrace{\text{measure of fit}}_{RSS(\theta)} + \;\lambda \cdot \underbrace{\text{measure of magnitude of coefficients}}_{\lambda \cdot \lVert \theta \rVert_1} \\ \\ \implies \ \min\; \lVert \Phi \theta - y \rVert_2^2 + \lambda \lVert \theta \rVert_1 \end{align*}\]-
\(\lambda\) is a tuning parameter = balance of fit and sparsity
-
Another equivalent forms of optimizations
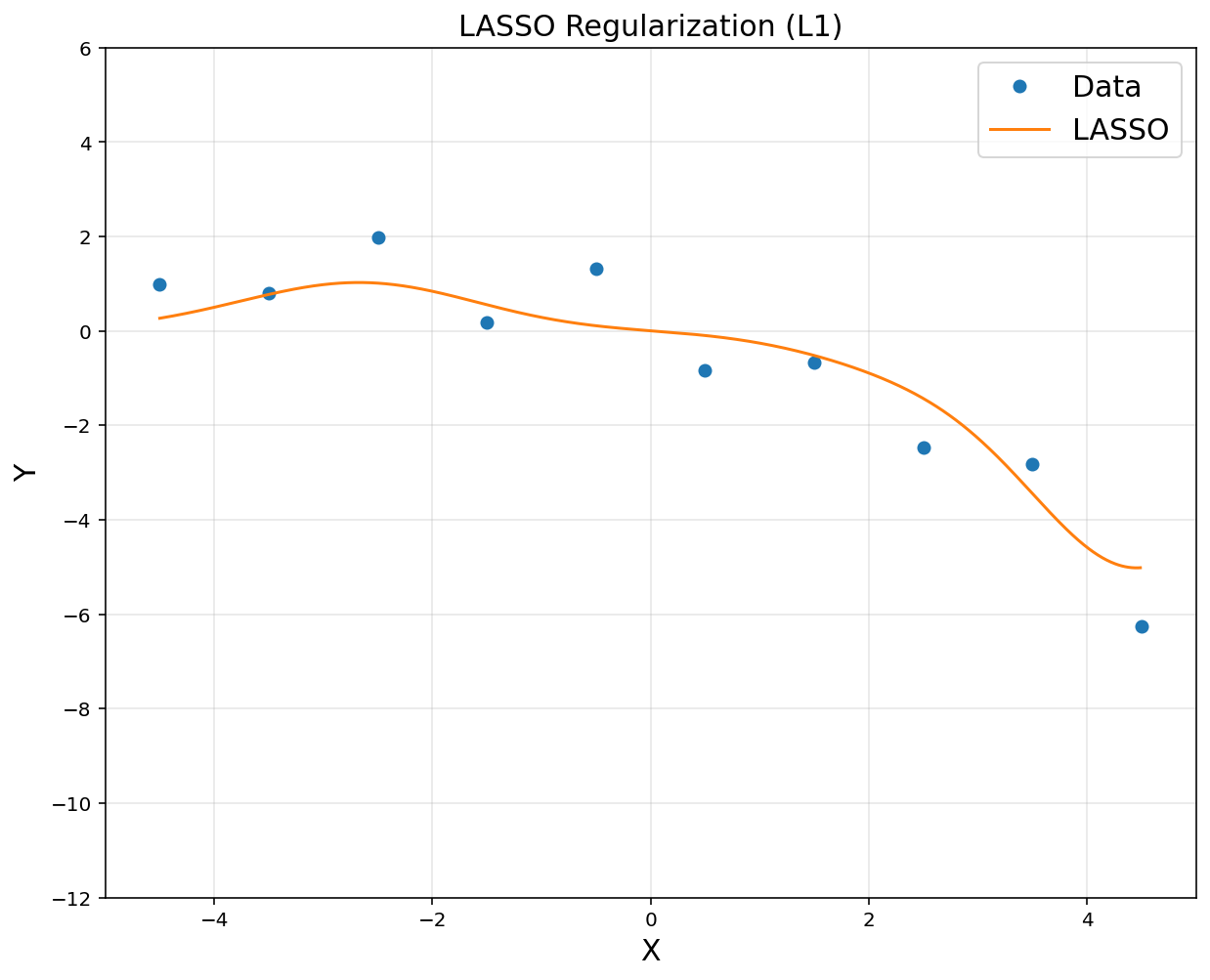
# LASSO regression
lamb = 2
theta = cvx.Variable([d, 1])
obj = cvx.Minimize(cvx.sum_squares(A@theta - y) + lamb*cvx.norm(theta, 1))
prob = cvx.Problem(obj).solve()
yp = rbfbasis@theta.value
plt.figure(figsize = (10, 8))
plt.title('LASSO Regularization (L1)', fontsize = 15)
plt.xlabel('X', fontsize = 15)
plt.ylabel('Y', fontsize = 15)
plt.plot(x, y, 'o', label = 'Data')
plt.plot(xp, yp, label = 'LASSO')
plt.axis([-5, 5, -12, 6])
plt.legend(fontsize = 15)
plt.grid(alpha = 0.3)
plt.show()
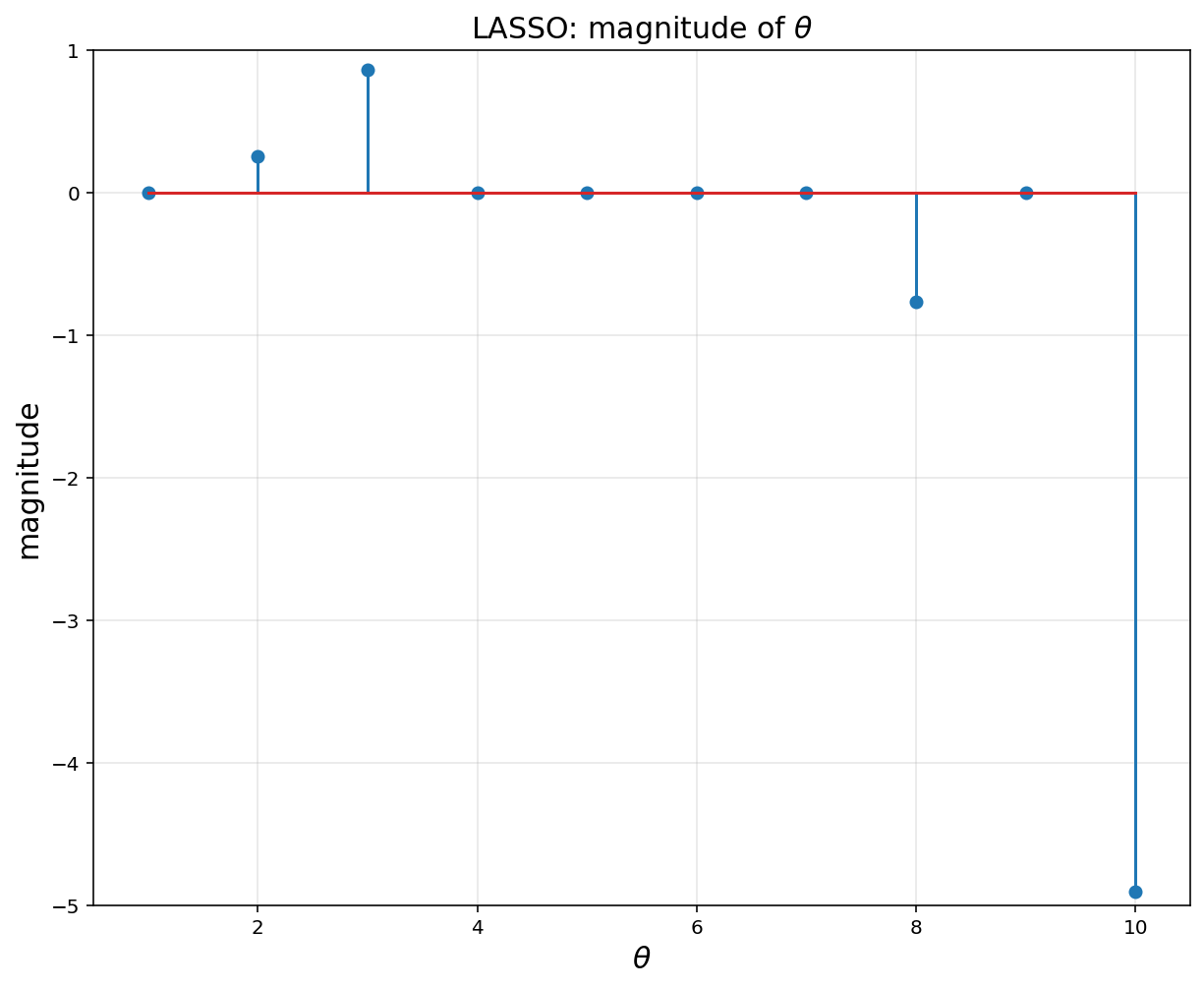
# Regulization (= Lasso nonlinear regression) encourages zero weights
plt.figure(figsize = (10, 8))
plt.title(r'LASSO: magnitude of $$\theta$$', fontsize = 15)
plt.xlabel(r'$$\theta$$', fontsize = 15)
plt.ylabel('magnitude', fontsize = 15)
plt.stem(np.arange(1,11), theta.value)
plt.xlim([0.5, 10.5])
plt.ylim([-5,1])
plt.grid(alpha = 0.3)
plt.show()
lamb = np.arange(0,3,0.01)
theta_record = []
for k in lamb:
theta = cvx.Variable([d, 1])
obj = cvx.Minimize(cvx.sum_squares(A@theta - y) + k*cvx.norm(theta, 1))
prob = cvx.Problem(obj).solve()
theta_record.append(np.ravel(theta.value))
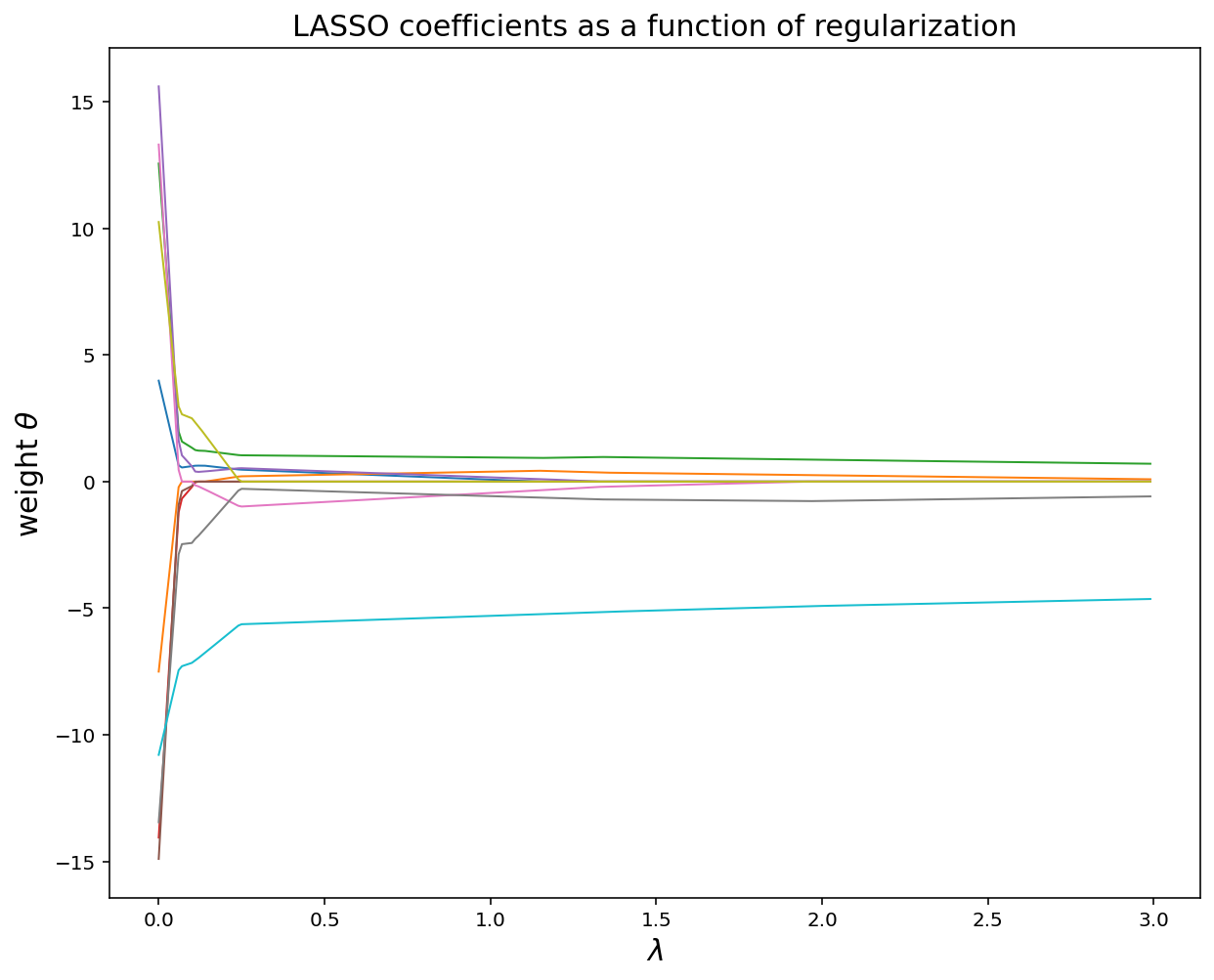
plt.figure(figsize = (10, 8))
plt.plot(lamb, theta_record, linewidth = 1)
plt.title('LASSO coefficients as a function of regularization', fontsize = 15)
plt.xlabel('$$\lambda$$', fontsize = 15)
plt.ylabel(r'weight $$\theta$$', fontsize = 15)
plt.show()
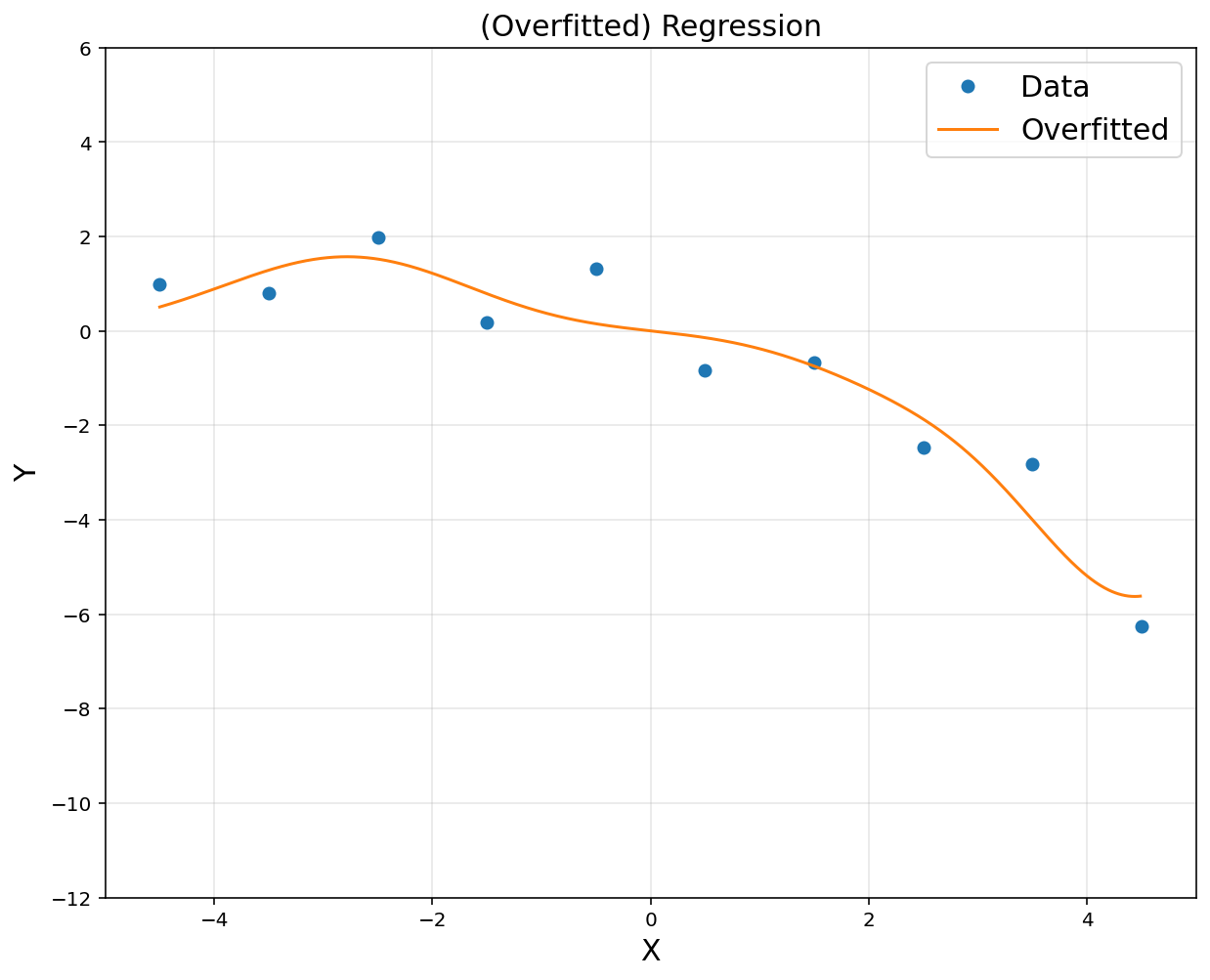
# reduced order model
# we will use only theta 2, 3, 8, 10
d = 4
u = np.array([-3.5, -2.5, 2.5, 4.5])
sigma = 1
A = np.hstack([np.exp(-(x-u[i])**2/(2*sigma**2)) for i in range(d)])
rbfbasis = np.hstack([np.exp(-(xp-u[i])**2/(2*sigma**2)) for i in range(d)])
A = np.asmatrix(A)
rbfbasis = np.asmatrix(rbfbasis)
theta = cvx.Variable([d, 1])
obj = cvx.Minimize(cvx.norm(A@theta-y, 2))
prob = cvx.Problem(obj).solve()
yp = rbfbasis@theta.value
plt.figure(figsize = (10, 8))
plt.plot(x, y, 'o', label = 'Data')
plt.plot(xp, yp, label = 'Overfitted')
plt.title('(Overfitted) Regression', fontsize = 15)
plt.xlabel('X', fontsize = 15)
plt.ylabel('Y', fontsize = 15)
plt.axis([-5, 5, -12, 6])
plt.legend(fontsize = 15)
plt.grid(alpha = 0.3)
plt.show()
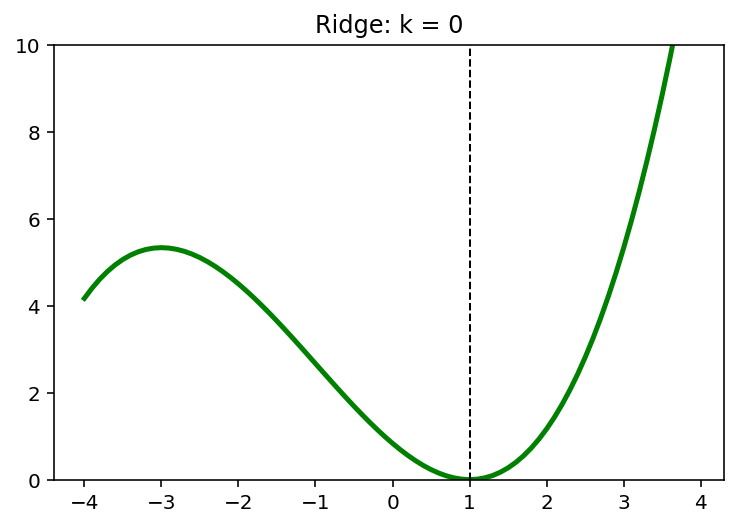
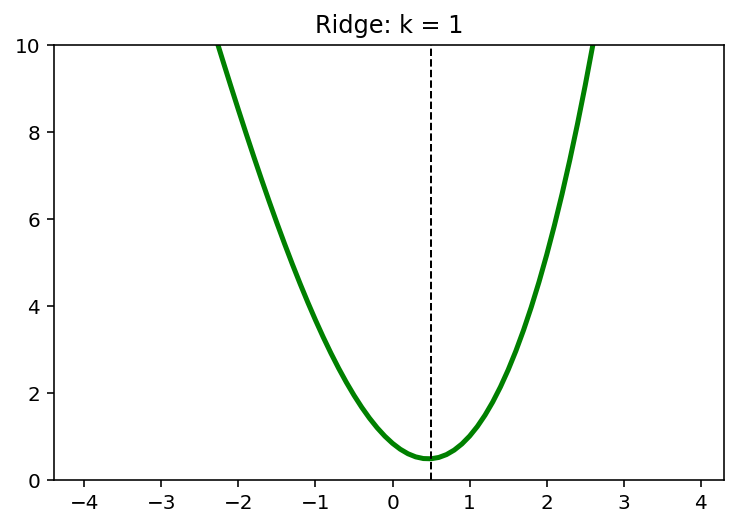
5. L2 and L1 Regularizers
\[\min \;(x-1)^2 + \frac{1}{6}(x-1)\]5.1. L2 Penality
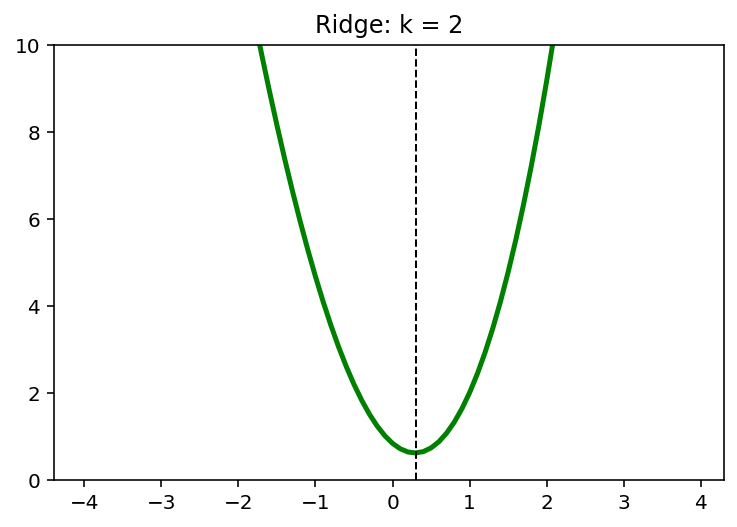
\[\min \; (x-1)^2 + \frac{1}{6}(x-1) + kx^2, \quad k=0,1,2,\cdots\]x = np.arange(-4,4,0.1)
k = 4
y = (x-1)**2 + 1/6*(x-1)**3 + k*x**2
x_star = x[np.argmin(y)]
print(x_star)
0.20000000000000373
plt.plot(x, y, 'g', linewidth = 2.5)
plt.axvline(x = x_star, color = 'k', linewidth = 1, linestyle = '--')
plt.ylim([0,10])
plt.show()
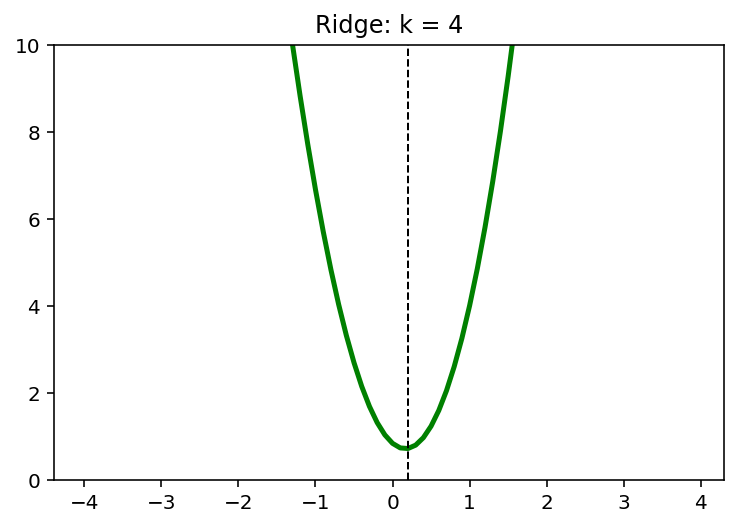
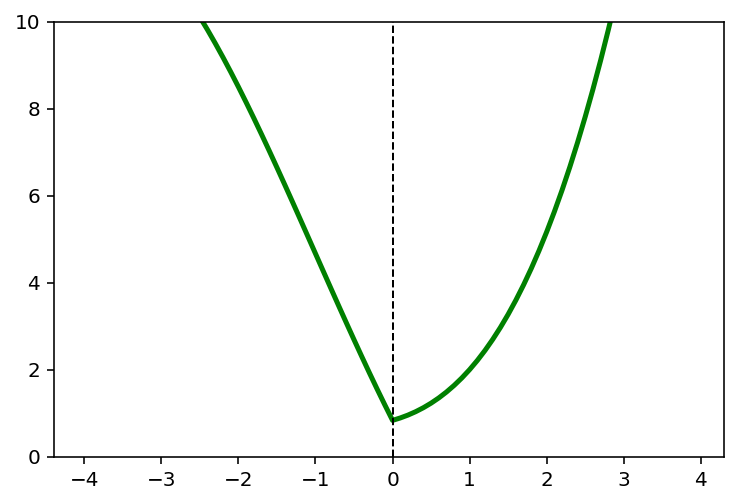
for k in [0,1,2,4]:
y = (x-1)**2 + 1/6*(x-1)**3 + k*x**2
x_star = x[np.argmin(y)]
plt.plot(x,y, 'g', linewidth = 2.5)
plt.axvline(x = x_star, color = 'k', linewidth = 1, linestyle = '--')
plt.ylim([0,10])
plt.title('Ridge: k = {}'.format(k))
plt.show()
5.2. L1 Penalty
\[\min \; (x-1)^2 + \frac{1}{6}(x-1) + k\lvert x \rvert, \quad k=0,1,2,\cdots\]x = np.arange(-4,4,0.1)
k = 2
y = (x-1)**2 + 1/6*(x-1)**3 + k*abs(x)
x_star = x[np.argmin(y)]
print(x_star)
3.552713678800501e-15 ```python plt.plot(x, y, 'g', linewidth = 2.5) plt.axvline(x = x_star, color = 'k', linewidth = 1, linestyle = '--') plt.ylim([0,10]) plt.show() ```
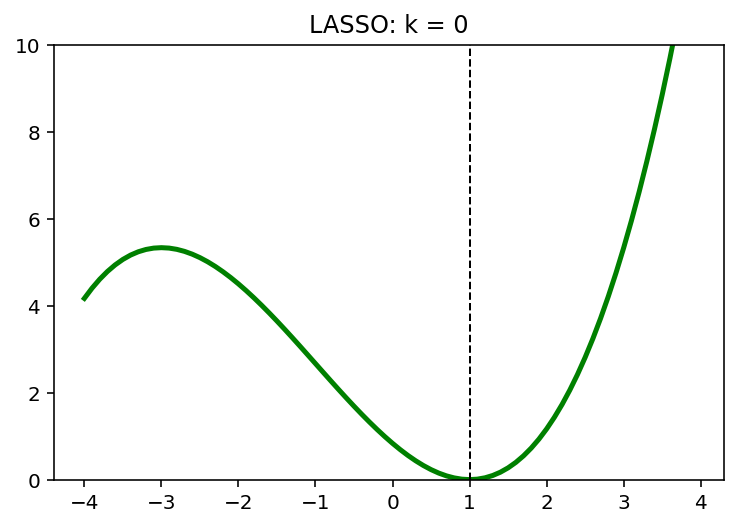
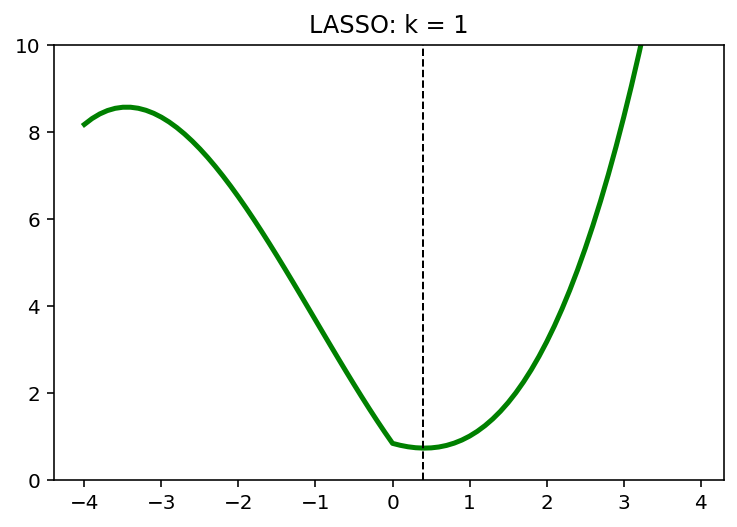
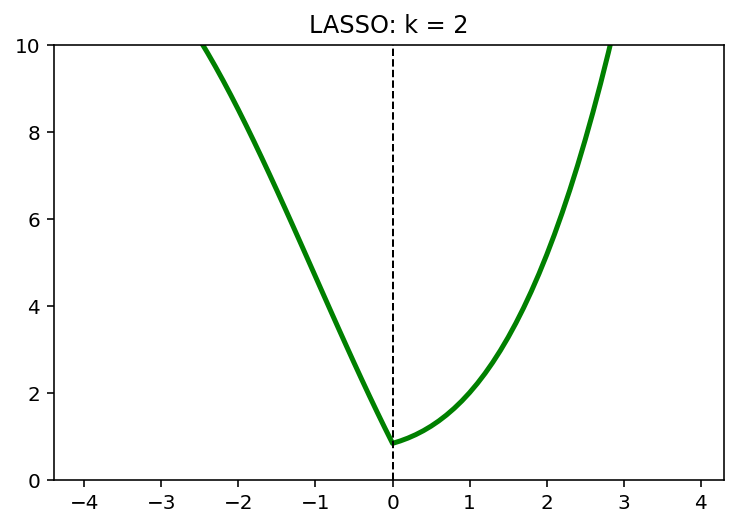
for k in [0,1,2]:
y = (x-1)**2 + 1/6*(x-1)**3 + k*abs(x)
x_star = x[np.argmin(y)]
plt.plot(x,y, 'g', linewidth = 2.5)
plt.axvline(x = x_star, color = 'k', linewidth = 1, linestyle = '--')
plt.ylim([0,10])
plt.title('LASSO: k = {}'.format(k))
plt.show()
6. Scikit-Learn
- RBF functions \(g_i(x)\) as basis
# 10 data points
n = 10
x = np.linspace(-4.5, 4.5, 10).reshape(-1, 1)
y = np.array([0.9819, 0.7973, 1.9737, 0.1838, 1.3180, -0.8361, -0.6591, -2.4701, -2.8122, -6.2512]).reshape(-1, 1)
d = 10
u = np.linspace(-4.5, 4.5, d)
sigma = 1
A = np.hstack([np.exp(-(x-u[i])**2/(2*sigma**2)) for i in range(d)])
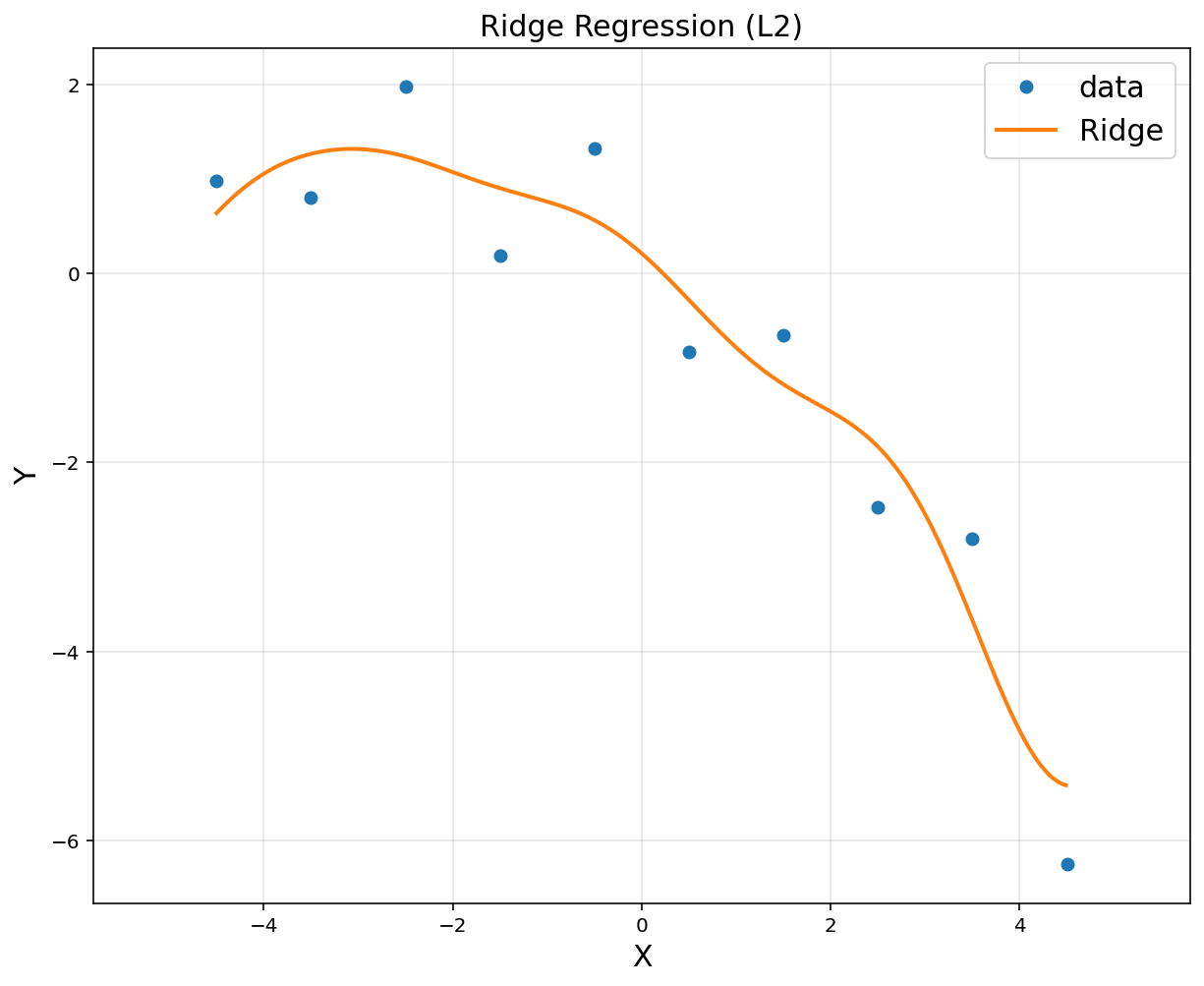
#from sklearn import linear_model
reg = linear_model.Ridge(alpha = 0.1)
reg.fit(A,y)
Ridge(alpha=0.1)
# to plot
xp = np.arange(-4.5, 4.5, 0.01).reshape(-1, 1)
Ap = np.hstack([np.exp(-(xp-u[i])**2/(2*sigma**2)) for i in range(d)])
plt.figure(figsize = (10, 8))
plt.plot(x, y, 'o', label = "data")
plt.plot(xp, reg.predict(Ap), linewidth = 2, label = "Ridge")
plt.title('Ridge Regression (L2)', fontsize = 15)
plt.xlabel('X', fontsize = 15)
plt.ylabel('Y', fontsize = 15)
plt.legend(fontsize = 15)
plt.axis('equal')
plt.grid(alpha = 0.3)
plt.show()
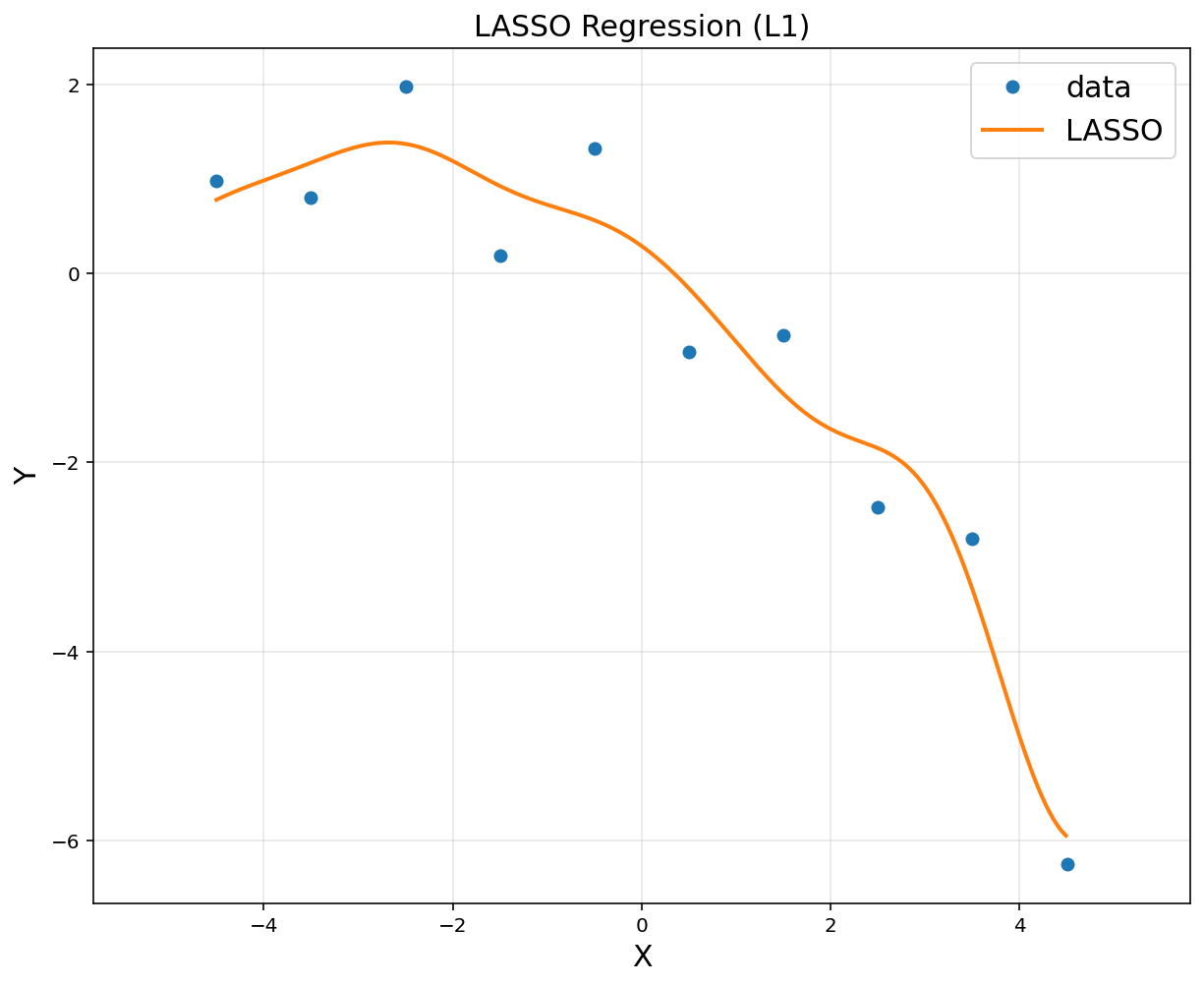
reg = linear_model.Lasso(alpha = 0.005)
reg.fit(A,y)
Lasso(alpha=0.005)
# to plot
xp = np.arange(-4.5, 4.5, 0.01).reshape(-1, 1)
Ap = np.hstack([np.exp(-(xp-u[i])**2/(2*sigma**2)) for i in range(d)])
plt.figure(figsize = (10, 8))
plt.plot(x, y, 'o', label = "data")
plt.plot(xp, reg.predict(Ap), linewidth = 2, label = "LASSO")
plt.title('LASSO Regression (L1)', fontsize = 15)
plt.xlabel('X', fontsize = 15)
plt.ylabel('Y', fontsize = 15)
plt.legend(fontsize = 15)
plt.axis('equal')
plt.grid(alpha = 0.3)
plt.show()

Leave a comment